New Disease-Causing Organisms in Alfalfa:
Aphanomyces Euteiches Race 2 and Phytoplasma
by R.D. Peters and C.R. Grau
Department of Plant Pathology
University of Wisconsin – Madison
Introduction
Alfalfa is the primary forage crop in Wisconsin and is a key element in the states dairy industry. The yield of new varieties is greater than that of Vernal and other older varieties due to genetic gains made by breeders over the years. Nevertheless, overall yields have declined steadily in Wisconsin during the past 30 years (Wiersma et al. 1997), even though efforts have been made to breed for resistance to a wide variety of major pathogens of alfalfa such as Verticillium, Phytophthora, and Aphanomyces. There are several possible explanations for this lack of progress including changing climatic and soil conditions and the difficulties inherent in dealing with such a genetically diverse crop. From a pathologist’s perspective, new disease-causing organisms or new strains of previously-described pathogens may also play a role in limiting yield gains for Wisconsin alfalfa growers. At the University of Wisconsin-Madison, we are conducting research on alfalfa diseases, particularly those caused by organisms that have not been studied previously with respect to their presence and influence on the alfalfa crop. What follows is a short summary of the biology and potential impact of two new pathogens: Aphanomyces euteiches Race 2 and phytoplasma.
Aphanomyces Root Rot
Aphanomyces root rot is caused by the pathogen Aphanomyces euteiches. It is most severe in flooded soil conditions and often associated with other root-rotting pathogens such as Phytophthora medicaginis (the pathogen causing Phytophthora root rot). Infection with Aphanomyces can result in the death of seedlings, but more often results in stunted, chlorotic (yellow) plants. Roots and hypocotyls develop light to dark brown lesions, but unlike other root-rot pathogens which cause seedling collapse, hypocotyls infected with Aphanomyces tend to remain rigid, resulting in stunted but upright seedlings (Grau 1990). Regrowth after harvest or winter dormancy is reduced in infected plants (Grau 1990).
Aphanomyces survives as oospores (sexual spores) in the soil or in infected plant tissues. Flooded conditions encourage oospores to germinate producing free-swimming zoospores (zoospores are also produced from germinating asexual primary spores). It is the zoospores that encyst on host roots causing infection to occur. Soil temperatures of 75-82°F are optimum for infection and the development of disease (Grau 1990).
Soils infested with Aphanomyces inoculum can be detected by planting bait plants into pots containing the field soil of interest and maintaining flooded conditions for 5 days. Plants with symptoms are then removed from the pots and root tissue from these plants is plated out onto a selective growth medium to obtain pure cultures of the pathogen (Pfender et al. 1984). The organism grows well on corn meal agar at room temperature, but must be transferred every month or so to maintain its viability (Grau 1990). Using a specialized technique (Mitchell and Yang 1966), pure cultures can be used to produce zoospores which can then be utilized to inoculate plant seedlings in the greenhouse or growth room to determine the resistance responses of germplasm to specific isolates of the pathogen.
Studies have noted that isolates of Aphanomyces tend to be most virulent (i.e. cause the most disease) on the crops from which they have been isolated (Malvick et al. 1998). In other words, isolates from alfalfa tend to cause more disease on alfalfa than on peas and vice versa. Exceptions do occur, and some isolates can cause severe disease on more than one crop (Malvick et al. 1998). The use of molecular tools has documented genotypic diversity among isolates of Aphanomyces obtained from peas (Malvick and Percich 1998) and significant genotypic diversity among isolates obtained from different host species (Malvick et al. 1998). This level of diversity (which indicates that there are genetic differences among populations of the pathogen; they are not all the same) in populations of Aphanomyces provides challenges for breeding programs, because specific resistances may not be active against all strains of the pathogen.
Fungicides containing metalaxyl (i.e. Apron) are not active against Aphanomyces and therefore, avoiding poorly drained soils and using resistant varieties are the main methods useful for control. WAPH-1 alfalfa germplasm was released by the Wisconsin Agricultural Experiment Station in 1989 (Grau 1992). This germplasm is resistant to isolates of Aphanomyces commonly found in Wisconsin (Race 1). Resistance to Race 1 of Aphanomyces has been widely incorporated into commercial alfalfa varieties (Malvick 1998). Around 1990, isolates that were highly virulent (i.e. caused significant disease) to breeding lines with resistance to Race 1 were recovered from soils collected in eastern and southern states. Similar isolates were also found in scattered locations in Wisconsin. Such isolates have since been coined “Race 2″ isolates and represent a new form of the pathogen. Race 2 isolates of Aphanomyces have now been found in Idaho, Maryland, Minnesota, Mississippi, North Carolina, Tennessee, Virginia, Iowa, Kentucky, and Wisconsin (Malvick 1998). Race 2 is therefore widely distributed in the United States. The distribution of Race 2 populations within a state is also important. In Wisconsin, southwestern counties seem to have a higher incidence of Race 2 isolates than other areas. The Lancaster Research Station is highly infested with Race 2 populations and most isolates from eastern Iowa are also Race 2. By contrast, fields at the Marshfield Research Station have a lower incidence of Race 2, although this incidence appears to be increasing.
Alfalfa breeding programs are developing lines that are resistant to Race 2 of Aphanomyces (Malvick 1998). Fortunately, these lines also appear to be highly resistant to Race 1 isolates as well. Race 2 resistant material would show the most pronounced effects where Race 2 populations of the pathogen predominate. However, even in regions where Race 1 populations predominate now, Race 2 may become more prevalent in the future if there is selection pressure by the crop (i.e. growing only Race 1 resistant varieties). The time frame for the development of resistant populations of the pathogen is unclear. However, the variability that is present in populations of Aphanomyces implies that breeders and growers must continue to be vigilant to meet the demands of controlling a changing pathogen population.
Phytoplasma
Phytoplasmas may best be described as bacteria that have lost their cell walls. They are only enclosed by a thin membrane which makes them very pliable and able to assume a variety of shapes (think of a balloon filled with water that can be squeezed into different shapes). They exist exclusively in the phloem (the carbohydrate-conducting tissue of the plant) and are transferred from plant to plant by insect vectors, primarily leafhoppers. Since their discovery in the late 1960s, phytoplasmas have been found in hundreds of different plant species and cause a variety of diseases. Symptoms of infection may include yellowing, stunted growth and slow decline (particularly in association with many tree species), and abnormal growth such as proliferation (an abnormally large number) of stems and buds and vegetative growth (such as leaves) from floral parts (phyllody). Although some strains of phytoplasma are found only on one crop, others, such as the organism causing aster yellows, can infect a wide range of host species.
One of the major difficulties of working with phytoplasma is the inability to grow the organism in culture. This makes detection more difficult. Traditional techniques that rely on plant symptoms to determine infection are not always reliable. An excellent collaboration has been established between our lab at U. of Wisconsin-Madison and Dr. Ing-Ming Lee, USDA, Beltsville, Maryland relative to phytoplasmas in soybean. This collaboration has been extended to cover alfalfa disease problems. Dr. Lee is a leading researcher in phytoplasma detection and taxonomy and has pioneered ground-breaking research in this area (Lee et al. 1998). Not only has Dr. Lee assisted in the establishment of phytoplasma detection and identification protocols at U. of Wisconsin-Madison, he has performed replicate experiments to confirm and validate results obtained in our laboratory.
A survey was conducted during September to November, 1998 to determine the incidence of phytoplasma in alfalfa plantings. Samples were obtained from fields near Arlington, Evansville, Marshfield, West Madison, Lancaster, Whitewater, Hancock and a grower field west of Madison. Samples consisted of growing tips from upper regions of plants. Samples were placed into labeled plastic bags and then placed in Styrofoam coolers for return to UW-Madison upon which samples were frozen at -20°C. Samples were processed by extraction of DNA using the protocol of (Zhang et al. 1998). This process separates the DNA (which contains the genes we are interested in) from the rest of the plant material. After the purified DNA was obtained, nested PCR (polymerase chain reaction) was carried out using two universal primer pairs according to the protocol of Gunderson and Lee (1996). The PCR process allows us to detect the DNA from only the phytoplasmas present in a sample and distinguishes phytoplasma DNA from all the other types of DNA present in the sample (such as plant, bacteria, and fungal DNA). This new technique (PCR) is costly and time-consuming, but allows us to detect organisms such as phytoplasmas in alfalfa (and other crops) which have probably been present in plantings for many years, but were very difficult to detect with traditional methods. For further classification of phytoplasmas, restriction enzyme digests were performed (the phytoplasma DNA was cut with enzymes) and comparison of RFLP (restriction fragment length polymorphisms) were made with known patterns described by Lee et al. (1998). In other words, after we cut the phytoplasma DNA with enzymes, the patterns that were produced were compared with patterns of other phytoplasmas from around the world to further identify what phytoplasmas we are dealing with in Wisconsin.
Results indicated that phytoplasmas or organisms closely related to phytoplasmas are widespread in alfalfa plantings in Wisconsin (see Table 1).
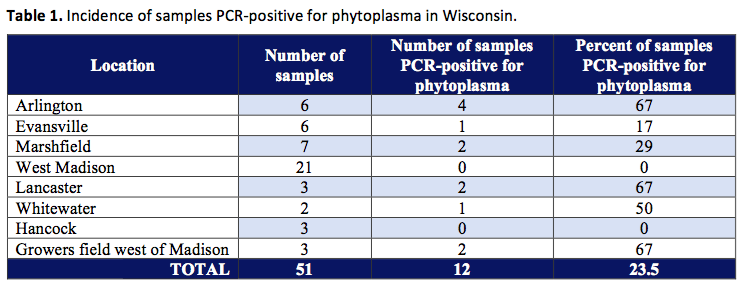
Restriction digest work (cutting with enzymes) on the PCR products (amplified DNA) of the 12 positive samples is continuing, but initial results have revealed the presence of phytoplasmas belonging to the aster yellows grouping as well as other organisms that are not currently recognized as phytoplasmas in the taxonomic system developed by Lee et al. (1998). This is the first report of the presence of the aster yellows organism in alfalfa. Aster yellows has been reported in other leguminous crops such as clover, broad bean, bur-clover (Medicago hispida), and soybean (McCoy et al. 1989, M.E. Lee, personal communication). The presence of clover proliferation strains is also likely based on its presence in sweet clover (McCoy et al. 1989) and in soybeans in Wisconsin (M.E. Lee, personal communication). More than one phytoplasma strain may also occur in a single plant (Alma et al. 1996, Bianco et al. 1993, Lee et al. 1995). In the current study, the presence of phytoplasma or related organisms was associated with plant samples displaying symptoms of purpling of the foliage, interveinal chlorosis (yellowing between plant veins), and leaf puckering. Purpling and chlorotic discoloration (yellowing) are symptoms commonly associated with infection by aster yellows phytoplasmas in other crops (Khurana et al. 1988, Zitter 1991). However, the presence of these symptoms did not always imply the presence of the pathogen, as PCR negative results were also obtained from plant samples showing these symptoms. More rigorous surveys, are required to assess the full extent of phytoplasma incidence in alfalfa plantings in Wisconsin.
Alfalfa is distinguished from other agricultural crops in having a perennial habit. In this way, it can be compared to many tree species that have been shown to harbor the phytoplasma organism for many seasons, resulting in chronic disease. Most our PCR positive alfalfa samples originated from plantings in their third or fourth year. This may indicate that the perennial nature of the crop predisposes it to phytoplasma infection by allowing a longer time-frame for transmission by leafhopper vectors and for the build-up of inoculum in host tissues. Forest researchers have noted that phytoplasmas are often concentrated in root tissues (Seemüller, 1988). Our research has also shown that PCR positive results can be obtained in basal stem, crown, and root tissues as late as December (sampling through the winter months will continue to determine if the pathogen can be detected in these tissues throughout the winter). Several plants obtained from the Arlington Research Station in early December, yielded root, crown, and basal stem tissue that was PCR positive with universal phytoplasma primers. It is probable that phytoplasmas can over winter in these tissues as it does in roots of tree species (Seemüller, 1988) and in perennial weeds.
Our discovery of PCR positive late season basal tissues has several potential epidemiological consequences. Current dogma states that leafhoppers blown into Wisconsin from the southern U.S. are largely responsible for transmission of phytoplasma and disease spread. However, the presence of the organism in over-wintered tissue would lead to the availability of inoculum in early spring which could then be transmitted by local leafhopper populations. The major vector involved in transmission is unclear, however the aster leafhopper (Macrosteles fascifrons) would be a prime candidate. Although the potato leafhopper is not strictly a phloem feeder, its sheer numbers and shotgun approach to feeding in alfalfa also makes it a candidate for transmission. Research is needed to determine the vector species involved in transmission and the timing of transmission events. We have a collection of insects obtained during the 1998 field season which is awaiting analysis for the presence of phytoplasma. We are also growing plants obtained from Wisconsin fields in the greenhouse to act as a source for transmission studies. We are hoping to begin establishment of leafhopper populations for these studies in January. Transmission from infected to non-infected hosts may also be possible using dodder (Cuscuta spp.) to act as a bridge between plants (dodder is a parasitic plant) or via direct grafting of host tissues (Chiykowski 1988). Since Kochs postulates (rules that must be fulfilled to prove that an organism causes a disease) are difficult to complete in a non-culturable organism such as phytoplasma, transmission electron microscopy will be utilized to confirm PCR data relative to the success and mechanisms of transmission (Chen et al. 1989). In addition, field trials established at the Agricultural Research Station in Lancaster include many alfalfa varieties (including glandular hair varieties) and chemical spray regimens which are being assessed for impact relative to leafhopper injury. Samples from these plots have also been gathered to determine phytoplasma incidence. To date, we have found samples that are PCR positive for phytoplasma in both leafhopper-susceptible and resistant varieties. It is possible that glandular-haired varieties, although effective for controlling potato leafhopper feeding, do not control other leafhopper species or insects that are involved in the spread of phytoplasmas. The effects of chemical sprays on various types of insect species and on the incidence of phytoplasmas also needs to be determined.
Alfalfa plants store carbohydrates (starches and sugars) in their roots and crowns. These carbohydrate reserves are used for regrowth in the spring and after each cutting. Carbohydrate reserves are replenished when the plant reaches 6 to 8 inches tall (Undersander et al. 1994). Management practices such as cutting too frequently can deplete carbohydrate reserves and lower stand persistence. Similarly, the presence of phytoplasma in the phloem of infected plants could affect the movement and available amount of carbohydrate reserves for the plant leading to reduced productivity and stand persistence. The presence of phytoplasma over several years in a field could be partially responsible for the observed decline that often occurs in alfalfa plantings over time. It is often noted that such decline is not associated with any of the traditional alfalfa pathogens and, in the past, has been blamed on variety genetics or environmental conditions. It is our hypothesis that phloem-limited phytoplasma are having a delayed yet significant impact on forage yield due to utilization and interruption with transport of carbohydrates. Such an impact would only be noticed over time. To this date, direct experimental evidence supporting this hypothesis is lacking however, it is interesting to note that the growers field from which samples were obtained west of Madison was showing serious decline in its second year. Symptoms of common pathogens were not apparent in these samples, yet they were PCR positive with the universal phytoplasma primers. Although cause and effect has not been proven, this observation provides impetus to perform further research to test this hypothesis. Aster yellows phytoplasmas have significant agronomic effects on a wide variety of crops including carrot and lettuce (Errampalli et al. 1991) and barley (Chiykowski 1991) and can cause significant losses in crops such as cucurbits, celery, onion, and legumes (Provvidenti 1996). In addition, phytoplasmas have been associated with slow decline of various shrubs and tree species (Lee et al. 1995, Sinclair 1995, Sinclair et al. 1990, Sinclair et al. 1994).
The chronic presence of phytoplasma in alfalfa would also have implications for other crops. Field observations imply that soybeans planted near alfalfa tend to display greater symptoms of phytoplasma infection than soybeans in other locations. In this manner, alfalfa could be acting as a reservoir host for infection of other crops. Phytoplasma presence in such a bridge host may not only have an agronomic effect on the reservoir host itself, but also on surrounding crops.
Research in our lab has also shown that alfalfa seed harvested from plants with low seed yield can produce plants (after planting in a growth chamber) that are PCR positive with universal phytoplasma primers (M.E. Lee, personal communication). Although more research is required, the potential for seed transmission of phytoplasma would not only have significant epidemiological and agronomic consequences, but would also challenge current scientific thinking which states that such transmission cannot occur. This research would therefore have significant basic and applied aspects. This result warrants further research into the role of seed transmission in the epidemiology of the phytoplasma/alfalfa pathosystem.
A major project has been proposed at the University of Wisconsin-Madison to determine the distribution, characterization, modes of transmission and impact of phytoplasma in alfalfa in Wisconsin. The principle investigators are Craig R. Grau (Plant Pathology), David B. Hogg (Entomology), John L. Wedberg (Entomology), Daniel J. Undersander (Agronomy), Jerry D. Doll (Agronomy), Rob Alleman (Graduate Student), and Rick D. Peters (Plant Pathology). Studies are proposed to determine the distribution of phytoplasma on a regional, individual field, and individual plant basis. Weeds will also be screened as potential sources of the pathogen, since they can act as reservoirs for subsequent crop infection. The initial cornerstone of any IPM program is correct diagnosis. Molecular tools will be utilized to accurately detect and characterize the organism in alfalfa. These tools should assist growers directly in the diagnosis of diseases caused by phytoplasmas and will aid alfalfa breeders in developing programs to breed for resistance to these pathogens. Insect collections will be made in each year of the study to determine the principle insect vectors (primarily assumed to be leafhoppers) responsible for spread. These studies will be linked with field studies addressing control practices such as chemical insect control and host resistance (glandular-haired varieties) to examine the effect of current IPM practices on phytoplasma infection and to develop an effective IPM program that takes phytoplasma infection into account. Other modes of transmission will be examined to determine, for example, the importance of seed transmission in the epidemiology of disease. Finally, infected plants produced via infected seed, via direct transmission in the greenhouse using grafting, dodder or insect transmission systems, or via naturally-occurring inoculum will be used in greenhouse and field studies to determine the impact of phytoplasma infection on the yield and quality of alfalfa.
Summary
Aphanomyces Race 2
- Populations of Race 2 of Aphanomyces are widespread in the U.S.A. and in Wisconsin.
- The prevalence of Race 2 populations in southwestern Wisconsin has limited the performance of Race 1 resistant alfalfa varieties in that region.
- Race 2 resistance, which also confers resistance to Race 1, is being incorporated into future alfalfa varieties.
Phytoplasma
- Phytoplasmas are bacteria-like pathogens that have not previously been associated with alfalfa in Wisconsin.
- Phytoplasmas belonging to the aster yellows group and other unrecognized groups have been detected in alfalfa fields in Wisconsin using new molecular tools (PCR).
- PCR is costly and time-consuming, but very sensitive and allows organisms (such as phytoplasmas) to be detected that were difficult to detect in the past.
- Symptoms of infection with phytoplasma include yellowing and purpling of foliage and stunting which are like symptoms caused by insect-feeding, nutritional disorders and climatic effects.
- Phytoplasmas are transmitted by insects (most likely leafhoppers) and possibly seed.
- We believe that the presence of phytoplasmas in the food transport tissues of the plant is interfering with the transport and use of carbohydrate reserves leading to lower yields and loss of persistence.
- A major project has been proposed at the University of Wisconsin-Madison to determine the distribution, characterization, modes of transmission and impact of phytoplasma in alfalfa in Wisconsin.
References
Alma, A., R.E., Davis, M. Vibio, A. Danielli, D. Bosco, A. Arzone, and A. Bertaccini. 1996. Mixed infection of grapevines in northern Italy by phytoplasmas including 16S rRNA RFLP subgroup 16SrI-B strains previously unreported in the host. Plant Dis. 80: 418-421.
Bianco, P.A., R.E. Davis, J.P. Prince, I.-M. Lee, D.E. Gunderson, A. Fortusini, and G. Belli. 1993. Double and single infections by aster yellows and elm yellows MLOs in grapevines with symptoms characteristic of flavescence doree. Riv. Pat. Veg., S.V. 3: 69-82.
Chen, T.A., J.D. Lei, and C.P. Lin. 1989. Detection and identification of plant and insect mollicutes. Pages 393-424 in The Mycoplasmas Volume V: Spiroplasmas, Acholeplasmas, and Mycoplasmas of Plants and Arthropods, R.F. Whitcomb and J.G. Tully, eds. Academic Press Inc., San Diego, CA. 653 p.
Chiykowski, L.N. 1988. Maintenance of yellows-type mycoplasmalike organisms. Pages 123-134 in Tree Mycoplasmas and Mycoplasma Diseases, C. Hiruki, ed., The University of Alberta Press, Edmonton, AB. 245 pp.
Chiykowski, L.N. 1991. Reaction of additional barley cultivars to two aster yellows strains. Can. Plant Dis. Surv. 71: 143-145.
Errampalli, D., J. Fletcher, and P.L. Claypool. 1991. Incidence of yellows in carrot and lettuce and characterization of mycoplasmalike organism isolates in Oklahoma. Plant Dis. 75: 579-584.
Grau, C.R. 1990. Aphanomyces root rot. Pages 10-11 in Compendium of Alfalfa Diseases: Second Edition, D.L. Stuteville and D.C. Erwin, eds. APS Press, St. Paul, MN.
Grau, C.R. 1992. Registration of WAPH-1 alfalfa germplasm with resistance to Aphanomyces root rot. Crop Sci. 32: 287-288.
Gunderson, D.E. and I.-M. Lee. 1996. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol. Medit. 35: 114-151.
Khurana, S.M.P., R.A. Singh, and D.M. Kaley. 1988. Mycoplasma-associated potato diseases and their control in India. Pages 285-316 in Mycoplasma Diseases of Crops: Basic and Applied Aspects, K. Maramorosch and S.P. Raychaudhuri, eds. Springer-Verlag, NY. 456 pp.
Lee, I.-M., D.E. Gunderson-Rindal, R.E. Davis, and I.M. Bartoszyk. 1998. Revised classification scheme of phytoplasmas based on RFLP analyses of 16S rRNA and ribosomal protein gene sequences. Plant Dis.: In press.
Lee, I.-M., A. Bertaccini, M. Vibio, and D.E. Gunderson. 1995. Detection of multiple phytoplasmas in perennial fruit trees with decline symptoms in Italy. Phytopathology 85: 728-735.
Malvick, D.K. 1998. Aphanomyces Race 2 is distributed widely in Wisconsin and the United States. Page 1 in The Haymaker: Summer 1998. W-L Research, Inc., Evansville, WI.
Malvick, D.K., C.R. Grau, and J.A. Percich. 1998. Characterization of Aphanomyces euteiches strains based on pathogenicity tests and random amplified polymorphic DNA analyses. Mycol. Res. 102: 465- 475.
Malvick, D.K. and J.A. Percich. 1998. Genotypic and pathogenic diversity among pea-infecting strains of Aphanomyces euteiches from the central and western United States. Phytopathology 88: 915-921.
McCoy, R.E., A. Caudwell, C.J. Chang, T.A.Chen, L.N. Chiykowski, M.T. Cousin, J.L. Dale, G.T.N. de Leeuw, D.A. Golino, K.J. Hackett, B.C. Kirkpatrick, R. Marwitz, H. Petzold, R.C. Sinha, M. Sugiura, R.F. Whitcomb, I.L. Yang, B.M. Zhu, and E. Seemuller. 1989. Plant diseases associated with mycoplasma-like organisms. Pages 545-640 in The Mycoplasmas Volume V: Spiroplasmas, Acholeplasmas, and Mycoplasmas of Plants and Arthropods, R.F. Whitcomb and J.G. Tully, eds. Academic Press Inc., San Diego, CA. 653 p.
Mitchell, J.E. and C.Y. Yang. 1966. Factors affecting growth and development of Aphanomyces euteiches. Phytopathology 56: 917-922.
Pfender, W.F., P.A. Delwiche, C.R. Grau, and D.J. Hagedorn. 1984. A medium to enhance recovery of Aphanomyces from infected plant tissue. Plant Dis. 68: 845-847.
Provvidenti, R. 1996. Disease caused by a phytoplasma: Aster yellows. Page 37 in Compendium of Cucurbit Diseases, T.A. Zitter, D.L. Hopkins, and C.E. Thomas, eds. APS Press, St. Paul, MN 87 pp.
Seemüller, E. 1988. Colonization patterns of mycoplasmalike organisms in trees affected by apple proliferation and pear decline. Pages 179-192 in Tree Mycoplasmas and Mycoplasma Diseases, C. Hiruki, ed., The University of Alberta Press, Edmonton, AB. 245 pp.
Sinclair, W.A. 1995. Epidemiology of a slow-decline phytoplasmal disease: ash yellows on old field sites in New York State. Phytopathology 85: 123-128.
Sinclair, W.A., H.M. Griffiths, and I.-M. Lee. 1994. Mycoplasmalike organisms as causes of slow growth and decline of trees and shrubs. J. Aboriculture 20: 176-189.
Sinclair, W.A., R.J. Iuli, A.T. Dyer, P.T. Marshall, J.A. Matteoni, C.R. Hibben, G.R. Stanosz, and B.S. Burns. 1990. Ash yellows: Geographic range and association with decline of white ash. Plant Dis. 74: 604-607.
Undersander, D., N. Martin, D. Cosgrove, K. Kelling, M. Schmidt, J. Wedberg, R. Becker, C. Grau, J. Doll, and M.E. Rice. 1994. Alfalfa Management Guide. NCR547 North Central Regional Extension Publication, American Society of Agronomy Inc., Crop Science Society of America Inc., Soil Science Society of America Inc. 51 pp.
Wiersma, D.W., D.J. Undersander, J.G. Lauer, and C.R. Grau. 1997. Lack of alfalfa yield progress in the Midwest. 1997 Central Alfalfa Improvement Conference Proceedings.
Zhang, Y.-p., J.K. Uyemoto, and B.C. Kirkpatrick. 1998. A small-scale procedure for extracting nucleic acids from woody plants infected with various phytopathogens for PCR assay. J. Vir. Meth. 71: 45-50.
Zitter, T.A. 1991. Diseases caused by mycoplasmalike organisms: Aster yellows. Page 43 in Compendium of Tomato Diseases, J.B. Jones, J.P. Jones, R.E. Stall, and T.A. Zitter, eds. APS Press, St. Paul, MN. 73 pp.
![]()